Vascular NO/cGMP-signalling
Various physiological functions such as smooth muscle cell relaxation and platelet aggregation are regulated by the gaseous messenger nitric oxide (NO) and cyclic guanosine monophosphate (cGMP). To investigate the molecular mechanisms involved in NO/cGMP signaling and its regulation under physiological and pathophysiological conditions, we use ex vivo tissue models (e.g. vasomotor function studies in organ baths, Langendorff perfusion of isolated hearts), 2D and 3D cell culture models (e.g. endothelial cells, smooth muscle cells) and enzymatic assays with purified enzymes including NO synthase (NOS), soluble guanylyl cyclase (sGC). The results of the research will contribute to a deeper understanding of the role of NO/cGMP as a regulator of blood pressure and cardiac function in health and disease. In an extended approach, NO/cGMP signaling in blood vessels and the heart will be investigated with a special focus on lipid and carbohydrate homeostasis.
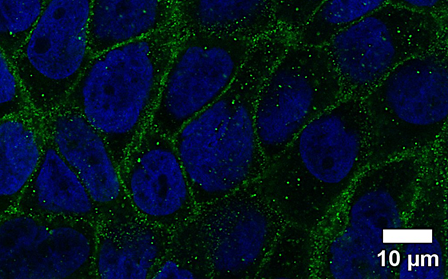
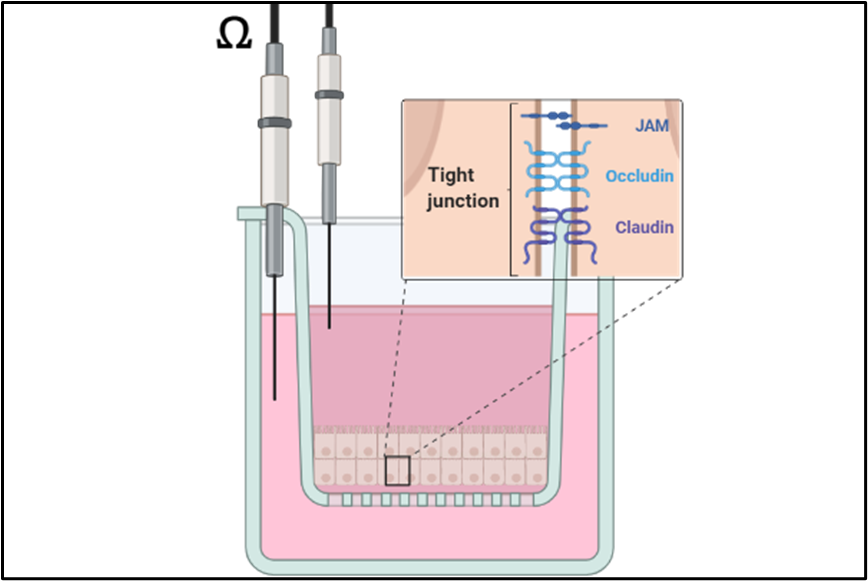
Epithelial Barrier of the Airways
The lungs are constantly exposed to a wide variety of environmental factors during ventilation. To protect the organism from noxious agents and pathogenic microorganisms, an intact barrier of the bronchial epithelium is required. Since this barrier is damaged by different factors (e.g. tobacco smoke, particulate matter, irritant gases), our research focuses on characterizing the mechanisms of various inhalation noxae and developing protective countermeasures (cooperation with the pharmacognosy department). To this end, mono- and co-cultures of human lung epithelial cells with vascular endothelial cells are placed on transwell inserts to serve as a model for a liquid-liquid or air-liquid interface. Gaseous substances can thus be tested directly or as an aqueous preparation under normoxic and hypoxic conditions. The integrity of the barrier is measured by functional assays (transepithelial electrical resistance, permeability assay) and characterized biochemically (Western blot, RT-qPCR) and by immunofluorescence microscopy.